What has become increasingly realised is that there is rather a lot of biological dark matter that we know simply nothing about at all and so the publication of a new tree of life (last Monday) that reveals quite a dramatically new picture of life is consequence of further insights. What has been happening is that a lot of new information was coming via genomic data and so this new tree of life represents the merger of over 1,000 uncultivated and previously unknown organisms, with the previously known sequences – in other words this new blending has yielded a dramatically expanded version of the tree of life.
Here it is below (the details of which are published here in Nature)
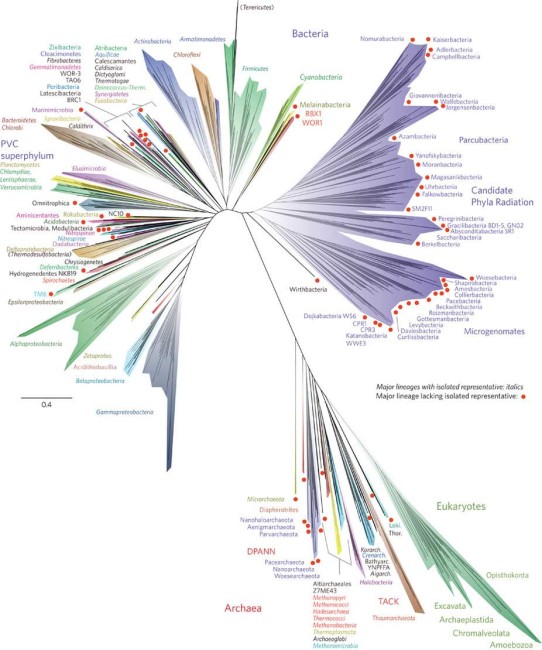
(You can click on it to get a larger version)
So what exactly is new here?
Up until now, representations of the tree of life tended to focus on the known well-classified diversity of life with an emphasis on eukaryotes. Now you might be wondering what the term “eukaryotes” means. Well that is basically any organism whose cells contain a nucleus and other organelles enclosed within membranes, hence includes not just us, but all animals, plants and also fungal organisms such as mushrooms (yes you are related to a Mushroom)
Now pay attention, and note that what this new tree of life reveals is that the eukaryote group is just a small part of the vast diversity of life. As they explain within their paper …
Here, we use new genomic data from over 1,000 uncultivated and little known organisms, together with published sequences, to infer a dramatically expanded version of the tree of life, with Bacteria, Archaea and Eukarya included.
.. and so what becomes a bit clearer is that the dominant form of life is Bacteria.
The challenge faced previously was that attempts to culture in the lab did not work for rather a lot of life and so that screamed out for a different way to measure and observe. The solution is the new bioinformatics. This involves obtaining samples of strands of DNA that is isolated directly from the environment, and then assembled, and that then yields complete or near-complete genome sequences without a reliance on cultivation.
Having got from this 1,011 newly reconstructed genomes they then merged that with genomes from public databases and created this new tree of life.
How do you construct a tree of life?
Did they just take the data and draw a picture to represent it all?
Actually no, the precise method they did use is described here in detail …
A total of 156 bootstrap replicates were conducted under the rapid bootstrapping algorithm, with 100 sampled to generate proportional support values. The full tree inference required 3,840 computational hours on the CIPRES supercomputer.
.. and so this is not just an artistic representation, it has rather a lot of serious analysis behind it and is the best fit for the data now available.
This is just the beginning
As things progress you can anticipate that our understanding of the tree of life will grow.
Clearly things may yet change, for example the current placement of Eukarya relative to Bacteria and Archaea is controversial and so more information might (or might not) lead to a revision of that.
One key observation, this really is the tip of the iceberg. The new data is just a sample of what is out there …
This study includes 1,011 organisms from lineages for which genomes were not previously available. The organisms were present in samples collected from a shallow aquifer system, a deep subsurface research site in Japan, a salt crust in the Atacama Desert, grassland meadow soil in northern California, a CO2-rich geyser system, and two dolphin mouths.
In other words, you can perhaps think of this as sticking your finger in the air to take a quick measurement. What is worth appreciating is that biologists are unable to culture and grow 89-99% of all living microorganisms, so there is a vast ocean of unknown biological dark matter still awaiting discovery.
We live in interesting times indeed.